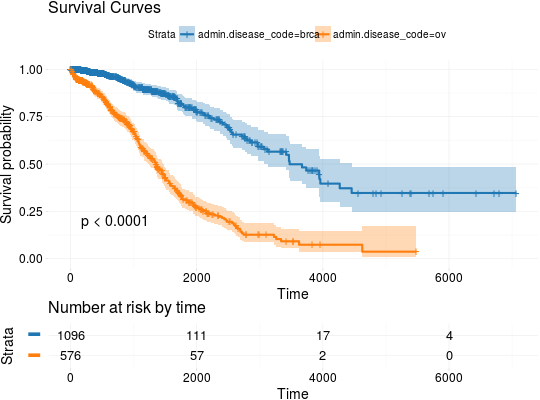
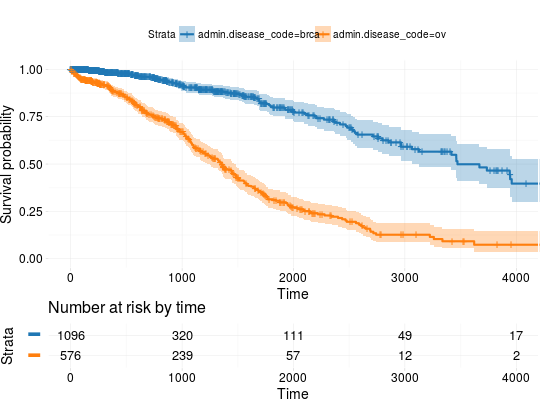
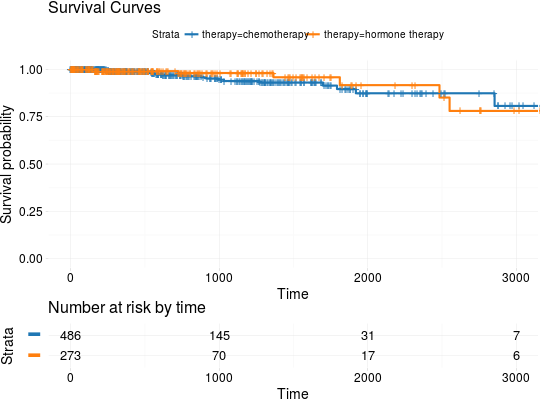
Extract Survival Information from Datasets Included in RTCGA.clinical and RTCGA.clinical.20160128 Packages
Extracts survival information from clicnial datasets from TCGA project.
survivalTCGA(..., extract.cols = NULL, extract.names = FALSE, barcode.name = "patient.bcr_patient_barcode", event.name = "patient.vital_status", days.to.followup.name = "patient.days_to_last_followup", days.to.death.name = "patient.days_to_death")
Arguments
- ...
- A data.frame or data.frames from TCGA study containing clinical informations. See clinical.
- extract.cols
- A character specifing the names of extra columns to be extracted with survival information.
- extract.names
- Logical, whether to extract names of passed data.frames in
.... - barcode.name
- A character with the name of
bcr_patient_barcodewhich differs between TCGA releases. By default is the name from the newest release datetail(checkTCGA('Dates'),1). - event.name
- A character with the name of
patient.vital_statuswhich differs between TCGA releases. By default is the name from the newest release datetail(checkTCGA('Dates'),1). - days.to.followup.name
- A character with the name of
patient.days_to_last_followupwhich differs between TCGA releases. By default is the name from the newest release datetail(checkTCGA('Dates'),1). - days.to.death.name
- A character with the name of
patient.days_to_deathwhich differs between TCGA releases. By default is the name from the newest release datetail(checkTCGA('Dates'),1).
Value
A data.frame containing information about times and censoring for specific bcr_patient_barcode.
The name passed in barcode.name is changed to bcr_patient_barcode.
Note
Input data.frames should contain columns patient.bcr_patient_barcode,
patient.vital_status, patient.days_to_last_followup, patient.days_to_death or theyir previous
equivalents.
It is recommended to use datasets from clinical.
Issues
If you have any problems, issues or think that something is missing or is not clear please post an issue on https://github.com/RTCGA/RTCGA/issues.
See also
RTCGA website http://rtcga.github.io/RTCGA/articles/Visualizations.html.
Other RTCGA: RTCGA-package,
boxplotTCGA, checkTCGA,
convertTCGA, createTCGA,
datasetsTCGA, downloadTCGA,
expressionsTCGA, heatmapTCGA,
infoTCGA, installTCGA,
kmTCGA, mutationsTCGA,
pcaTCGA, readTCGA,
theme_RTCGA
Examples
## Extracting Survival Data library(RTCGA.clinical) survivalTCGA(BRCA.clinical, OV.clinical, extract.cols = "admin.disease_code") -> BRCAOV.survInfo ## Kaplan-Meier Survival Curves kmTCGA(BRCAOV.survInfo, explanatory.names = "admin.disease_code", pval = TRUE)# first munge data, then extract survival info library(dplyr) BRCA.clinical %>% filter(patient.drugs.drug.therapy_types.therapy_type %in% c("chemotherapy", "hormone therapy")) %>% rename(therapy = patient.drugs.drug.therapy_types.therapy_type) %>% survivalTCGA(extract.cols = c("therapy")) -> BRCA.survInfo.chemo # first extract survival info, then munge data survivalTCGA(BRCA.clinical, extract.cols = c("patient.drugs.drug.therapy_types.therapy_type")) %>% filter(patient.drugs.drug.therapy_types.therapy_type %in% c("chemotherapy", "hormone therapy")) %>% rename(therapy = patient.drugs.drug.therapy_types.therapy_type) -> BRCA.survInfo.chemo kmTCGA(BRCA.survInfo.chemo, explanatory.names = "therapy", xlim = c(0, 3000), conf.int = FALSE)